Difference between revisions of "Research"
MarcRiedel (talk | contribs) |
|||
| (44 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
"''You see things; and you say, 'Why?' But I dream things that never were; and I say, 'Why not?'" | "''You see things; and you say, 'Why?' But I dream things that never were; and I say, 'Why not?'"'' | ||
''––'' '''George Bernard Shaw''' (1856 –1950) | |||
== | Our research spans different disciplines ranging from digital circuit design, to algorithms, to mathematics, to synthetic biology. It tends to be ''inductive'' (as opposed to ''deductive'') and ''conceptual'' (as opposed to ''applied''). A recurring theme is building systems that compute in novel or unexpected ways with new and emerging technologies. | ||
==Storing Data with Molecules== | |||
''All new ideas pass through three stages:'' | |||
# ''It can't be done.'' | |||
# ''It probably can be done, but it's not worth doing.'' | |||
# ''I knew it was a good idea all along!'' | |||
''––'''''Arthur C. Clarke (1917–2008)''' | |||
Ever since Watson and Crick first described the molecular structure of DNA, its information-bearing potential has been apparent. With each nucleotide in the sequence drawn from the four-valued alphabet of {A, T , C, G}, a molecule of DNA with ''n'' nucleotides stores ''2n'' bits of data. | |||
* Could we store data for our computer systems in DNA? ''"Can't be done '''–''''' ''too hard."'' | |||
* Is it worth doing? ''"Definitely not. It will never work as well as our hard drives do."'' | |||
* But one can store so much data so efficiently! "''I knew it was a good idea all along!''" | |||
<br> | |||
{| | |||
| | |||
{| style="background:#F0E68C" | |||
|- valign="top" | |||
| width="100" |'''title''': | |||
| width="500" |[http://mriedel.ece.umn.edu/wiki/images/8/8f/Manicka_Stephan_Chari_Mendonsa_Okubo_Stolzberg-Schray_Reddy_Riedel_Automated_Routing_of_Droplets_for_DNA_Storage_on_a_Digital_Microfluidics_Platform.pdf Automated Routing of Droplets for DNA Storage on a Digital Microfluidics Platform] | |||
|- valign="top" | |||
|'''authors''': | |||
|[http://mriedel.ece.umn.edu/wiki/index.php/Ajay_Manicka Ajay Manicka], Andrew Stephan, Sriram Chari, Gemma Mendonsa, | |||
Peyton Okubo, John Stolzberg-Schray, Anil Reddy, and [http://mriedel.ece.umn.edu/wiki/index.php/Marc_Riedel Marc Riedel] | |||
|- valign="top" | |||
|'''appeared in''': | |||
|[https://pubs.rsc.org/en/content/articlepdf/2023/DD/D3DD00083D?page=search Royal Society of Chemistry – Digital Discovery], Vol. 2, pp. 1436–1451, 2023 | |||
|} | |||
| width="70" align="center" |<span class="plainlinks"> [https://mriedel.ece.umn.edu/wiki/images/8/8f/Manicka_Stephan_Chari_Mendonsa_Okubo_Stolzberg-Schray_Reddy_Riedel_Automated_Routing_of_Droplets_for_DNA_Storage_on_a_Digital_Microfluidics_Platform.pdf http://mriedel.ece.umn.edu/wiki/images/0/04/Pdf.jpg]</span> | |||
[http://mriedel.ece.umn.edu/wiki/images/8/8f/Manicka_Stephan_Chari_Mendonsa_Okubo_Stolzberg-Schray_Reddy_Riedel_Automated_Routing_of_Droplets_for_DNA_Storage_on_a_Digital_Microfluidics_Platform.pdf Paper] | |||
| width="70" align="center" |<span class="plainlinks">[https://mriedel.ece.umn.edu/wiki/images/1/11/Dna-storage-and-computing.pptx http://mriedel.ece.umn.edu/wiki/images/3/36/Ppt.jpg]</span> | |||
[https://mriedel.ece.umn.edu/wiki/images/1/11/Dna-storage-and-computing.pptx Slides] | |||
|} | |||
<br> | |||
[[File:Dna-computing-dalle.jpg|center|thumb|Storing data in DNA.]] | |||
<br> | |||
==Computing with Molecules== | |||
"''Biology is the most powerful technology ever created. DNA is software, protein are hardware, cells are factories''." | |||
––'''Arvind Gupta (1953– )''' | |||
Computing has escaped! It has gone from desktops and data centers into the wild. Embedded microcontrollers – found in our gadgets, our buildings, and even our bodies – are transforming our lives. And yet, there are limits to where silicon can go and where it can compute effectively. It is a foreign object that requires a electrical power source. | |||
We are studying novel types of computing systems that are not foreign, but rather an integral part of their physical and chemical environments: systems that compute ''directly'' with molecules. A simple but radical idea: compute with '''acids''' and '''bases'''. An acidic solution corresponds to a "1" and a basic solution to "0". | |||
<br> | |||
|| | {| | ||
| | | | ||
{| style="background:#F0E68C" | |||
|- valign="top" | |||
| width="100" |'''title''': | |||
| width="500" |[http://mriedel.ece.umn.edu/wiki/images/0/02/Agiza_Oakley_Rosenstein_Rubenstein_Kim_Riedel_Reda_Digital_Circuits_and_Neural_Networks_Based_on_Acid-Base_Chemistry_Implemented_by_Robotic_Fluid_Handling.pdf Digital Circuits and Neural Networks Based on Acid-Base Chemistry Implemented by Robotic Fluid Handling] | |||
|- valign="top" | |||
|'''authors''': | |||
|[https://cs.brown.edu/people/grad/aagiza/ Ahmed Agiza], [https://www.linkedin.com/in/kady-oakley-5b05a75b Kady Oakley], [http://rosenstein.engin.brown.edu/ Jacob Rosenstein], [https://rubenstein.group/authors/brenda-rubenstein/ Brenda Rubenstein], | |||
[https://sites.brown.edu/kim-chemistry/eunsuk-kim/ Eunsuk Kim], [http://mriedel.ece.umn.edu/wiki/index.php/Marc_Riedel Marc Riedel], and [https://vivo.brown.edu/display/sreda Sherief Reda] | |||
|- valign="top" | |||
|'''appeared in''': | |||
|[https://www.nature.com/articles/s41467-023-36206-8 '''Nature Communications'''], Vol. 14, No. 496, 2023 | |||
|} | |||
|} | |} | ||
<br> | |||
We have developed | [[File:Computing-with-chemistry.jpg|center|thumb|450x450px|Computing with Acids and Bases]] | ||
<br> | |||
It's more complex that acids and bases, but DNA is a terrific chassis for computing. We have developed "'''CS 101'''" algorithms with DNA: ''Sorting, Shifting'' and ''Searching'': | |||
{| | |||
| | |||
{| style="background:#F0E68C" | |||
|- valign="top" | |||
| width="100" | '''title''': | |||
| width="500" | [https://mriedel.ece.umn.edu/wiki/images/b/b2/Solanki-chen-riedel-parallel-pairwise-operations-on-data-stored-in-dna-sorting-xor-shifting-and-searching.pdf Parallel Pairwise Operations on Data Stored in DNA: Sorting, XOR, Shifting, and Searching] | |||
|- valign="top" | |||
| '''authors''': | |||
| [[Arnav Solanki]], [[Tonglin Chen]], and [[Marc Riedel]] | |||
|- valign="top" | |||
| '''appeared in''': | |||
| [https://link.springer.com/article/10.1007/s11047-023-09964-z Natural Computing], 2023 | |||
|- valign="top" | |||
| '''presented at''': | |||
| [https://drops.dagstuhl.de/opus/portals/lipics/index.php?semnr=16207 International Conference on DNA Computing and Molecular Programming], 2021 | |||
|} | |||
| width="70" align="center" | | |||
<span class="plainlinks">[https://mriedel.ece.umn.edu/wiki/images/b/b2/Solanki-chen-riedel-parallel-pairwise-operations-on-data-stored-in-dna-sorting-xor-shifting-and-searching.pdf http://mriedel.ece.umn.edu/wiki/images/0/04/Pdf.jpg]</span> | |||
<br>[https://mriedel.ece.umn.edu/wiki/images/b/b2/Solanki-chen-riedel-parallel-pairwise-operations-on-data-stored-in-dna-sorting-xor-shifting-and-searching.pdf Paper] | |||
| width="70" align="center" | | |||
<span class="plainlinks">[http://mriedel.ece.umn.edu/wiki/images/d/d5/DNA27_Presentation.pdf http://mriedel.ece.umn.edu/wiki/images/3/36/Ppt.jpg]</span> | |||
<br> [http://mriedel.ece.umn.edu/wiki/images/d/d5/DNA27_Presentation.pdf Slides] | |||
|} | |||
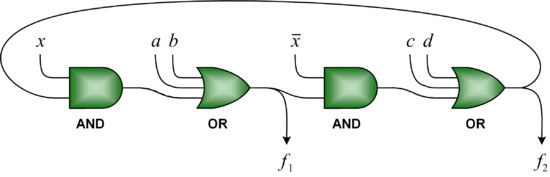
Based on a bistable mechanism for representing bits, we have implemented logic gates such '''AND''', '''OR''', and '''XOR gates''', as well as sequential components such as '''latches''' and '''flip-flops''' with DNA. Using these components, we have built full-fledged digital circuits such as a ''binary counters'' and ''linear feedback shift registers''. | |||
{| | {| | ||
| | | | ||
| Line 26: | Line 103: | ||
|- valign="top" | |- valign="top" | ||
| width="100" | '''title''': | | width="100" | '''title''': | ||
| width="500" | [[Media: | | width="500" | [[Media:Jiang_Riedel_Parhi_Digital_Logic_with_Molecular_Reactions.pdf | Digital Logic with Molecular Reactions]] | ||
|- valign="top" | |- valign="top" | ||
| '''authors''': | | '''authors''': | ||
| [[ | | [[Hua Jiang]], [[Marc Riedel]], [http://www.ece.umn.edu/users/parhi Keshab Parhi] | ||
|- valign="top" | |- valign="top" | ||
| '''presented at''': | |||
| [http://www.iccad.com The International Conference on Computer-Aided Design], San Jose, CA, 2013. | |||
|} | |} | ||
| align="center" width="70" | | | align="center" width="70" | | ||
<span class="plainlinks"> | <span class="plainlinks">[http://cctbio.ece.umn.edu/wiki/images/c/cf/Jiang_Riedel_Parhi_Digital_Logic_with_Molecular_Reactions.pdf http://mriedel.ece.umn.edu/wiki/images/0/04/Pdf.jpg]</span> | ||
[http:// | <br>[[Media:Jiang_Riedel_Parhi_Digital_Logic_with_Molecular_Reactions.pdf | Paper]] | ||
<br> | |||
[[Media: | |||
|} | |} | ||
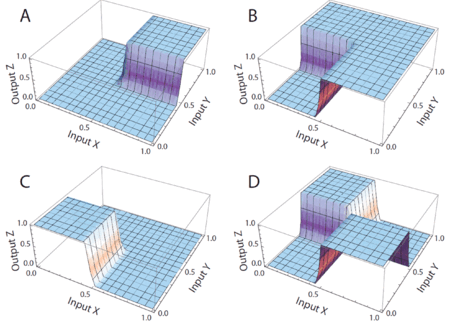
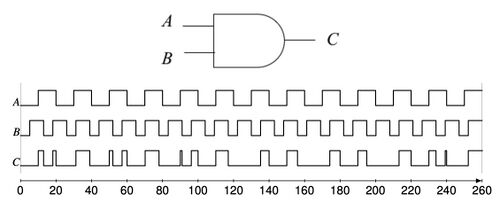
[[Image:dna-logic-gates.gif|center|thumb|450px| Simulations of DNA implementation of logic gates. The input signals are molecular concentrations X and Y; the output signal is a molecular concentration Z. (A) AND gate. (B) OR gate. (C) NOR gate. (D) XOR gate.]] | |||
Also, we have performed signal processing including operations such as '''filtering''' and '''fast-fourier transforms (FFTs)''' with DNA. | |||
{| | {| | ||
| | | | ||
| Line 49: | Line 124: | ||
|- valign="top" | |- valign="top" | ||
| width="100" | '''title''': | | width="100" | '''title''': | ||
| width="500" | [[Media: | | width="500" | [[Media:Jiang_Salehi_Riedel_Parhi_Discrete-Time_Signal_Processing_with_DNA.pdf | Discrete-Time Signal Processing with DNA]] | ||
|- valign="top" | |- valign="top" | ||
| '''authors''': | | '''authors''': | ||
| [http://www.ece.umn.edu/ | | [[Hua Jiang]], Ahmed Salehi, [[Marc Riedel]] and [http://www.ece.umn.edu/users/parhi Keshab Parhi] | ||
|- valign="top" | |- valign="top" | ||
| '''appeared in''': | | '''appeared in''': | ||
| [http:// | | [http://pubs.acs.org/doi/abs/10.1021/sb300087n ACS Synthetic Biology], Vol. 2 no. 5, pp. 245–254, 2013.<br>[[Media:Jiang_Salehi_Riedel_Parhi_Discrete-Time_Signal_Processing_with_DNA_Appendix.pdf | Supplementary Information: List of Reactions]] | ||
|- valign="top" | |- valign="top" | ||
| '''appeared in''': | |||
| [http://www.computer.org/portal/web/dt IEEE Design & Test of Computers], Vol. 29, No. 3, pp. 21–31, 2012. | |||
|- valign="top" | |||
| '''presented at''': | |||
| [http://www.iccad.com/ IEEE/ACM International Conference on Computer-Aided Design],<br> San Jose, CA, 2010. | |||
|- valign="top" | |||
| '''presented at''': | | '''presented at''': | ||
| [http:// | | [http://www.wikicfp.com/cfp/servlet/event.showcfp?eventid=7703©ownerid=8242 IEEE Workshop on Signal Processing Systems], San Francisco, 2010 | ||
|} | |} | ||
| align="center" width="70" | | | align="center" width="70" | | ||
<span class="plainlinks"> | <span class="plainlinks"> | ||
[http:// | [http://mriedel.ece.umn.edu/wiki/images/3/36/Jiang_Salehi_Riedel_Parhi_Discrete-Time_Signal_Processing_with_DNA.pdf http://mriedel.ece.umn.edu/wiki/images/0/04/Pdf.jpg]</span> | ||
<br> [http://mriedel.ece.umn.edu/wiki/images/3/36/Jiang_Salehi_Riedel_Parhi_Discrete-Time_Signal_Processing_with_DNA.pdf Paper] | |||
| align="center" width="70" | | |||
<span class="plainlinks"> | |||
[http://mriedel.ece.umn.edu/wiki/images/2/2a/Jiang_Kharam_Riedel_Parhi_Digital_Signal_Processing_with_Biomolecular_Reactions.pptx http://mriedel.ece.umn.edu/wiki/images/3/36/Ppt.jpg]</span> | |||
<br> [http://mriedel.ece.umn.edu/wiki/images/2/2a/Jiang_Kharam_Riedel_Parhi_Digital_Signal_Processing_with_Biomolecular_Reactions.pptx Slides] | |||
|} | |||
<br> | <br> | ||
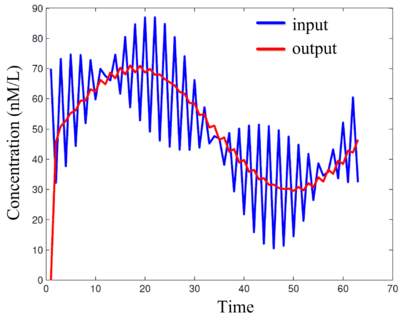
[[Image:moving-average-filter-simulation.gif|center|thumb|400px|Simulations of DNA implementation of a '''moving-average FIR filter'''. This filter removes the high-frequency component from an input signal, producing an output signal consisting of only the low-frequency component. Here the "signals" are molecular concentrations.]] | |||
Please see our "[[Papers,_Theses,_and_Presentations |Publications]]" page for more of our papers on these topics. | |||
==Computational Immunology== | |||
“''Physics is the study of the simple things in the Universe. Biology is the study of the complex ones.'' ” | |||
–– '''Richard Dawkins''' (1941– ) | |||
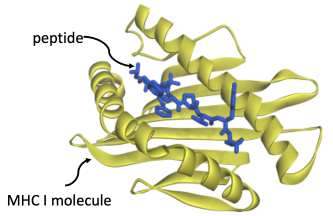
We are studying a problem that computer science currently judges to be very difficult: '''''predicting cellular immunity'''''. It centers on the question of how strongly molecules binds to one another. The molecules in question are ''peptides'' – fragments of proteins from a virus – binding to cell-surface receptors. A peptide will only bind if it fits like a key into a lock. | |||
{| align="center" | |||
|| | |||
[[Image:Peptide1.PNG|center|thumb|500px|A peptide (in blue) bound to a MHC Class I protein (in yellow).]] | |||
|| | |||
|| | |||
|} | |||
The binding is a critical step in a critical component of the immune system: it allows circulating '''T-cells to kill off infected cells.''' If this mechanism succeeds, an infection is stopped in its tracks. If it fails, then infected cells become factories for reproducing copies of the virus; full-blown disease results. Given a novel pathogen, such as SARS-Cov-2, predicting whether the immune system of an individual will do its job at fighting off the disease comes down to predicting how well the viral peptides bind to the cell-surface receptors of that person. We are tackling the problem with cloud computing resources, donated by [https://blogs.oracle.com/research/post/announcing-inaugural-cohort-oracle-research-fellows Oracle]. | |||
{| | {| | ||
| | | | ||
| Line 79: | Line 178: | ||
|- valign="top" | |- valign="top" | ||
| width="100" | '''title''': | | width="100" | '''title''': | ||
| width="500" | [ | | width="500" | [https://mriedel.ece.umn.edu/wiki/images/2/2d/Atkins_Solanki_Vasmatzis_Cornette_Riedel_Geographically_Biased_Composition_of_NetMHCpan_Training_Datasets_and_Evaluation_of_MHC-Peptide_Binding_Prediction_Accuracy_on_Novel_Alleles.pdf Geographically Biased Composition of NetMHCpan Training Datasets and Evaluation of MHC-Peptide] | ||
|- valign="top" | |- valign="top" | ||
| '''authors''': | | '''authors''': | ||
| | | Thomas Atkins, [[Arnav Solanki]], [https://www.mayo.edu/research/faculty/vasmatzis-george-ph-d/bio-00027811 George Vasmatzis], [https://orion.math.iastate.edu/cornette/ James Cornette], and [[Marc Riedel]] | ||
|- valign="top" | |- valign="top" | ||
| ''' | | '''appeared in''': | ||
| [ | | [https://www.frontiersin.org/journals/immunology Frontiers in Immunology], Vol. 14, 2023 | ||
|} | |} | ||
| align="center" width="70" | | | align="center" width="70" | | ||
<span class="plainlinks"> | <span class="plainlinks"> | ||
[ | [https://www.frontiersin.org/articles/10.3389/fimmu.2023.1288105/full http://mriedel.ece.umn.edu/wiki/images/0/04/Pdf.jpg]</span> | ||
<br> | <br> | ||
[ | [https://www.frontiersin.org/articles/10.3389/fimmu.2023.1288105/full Paper] | ||
|} | |} | ||
{| | {| | ||
| | | | ||
| Line 109: | Line 198: | ||
|- valign="top" | |- valign="top" | ||
| width="100" | '''title''': | | width="100" | '''title''': | ||
| width="500" | [[Media: | | width="500" | [[Media:Evasive_Spike_Variants_Elucidate_the_Preservation_of_T_Cell_Immune_Response_to_the_SARS-CoV-2_Omicron_Variant.pdf|Evasive Spike Variants Elucidate the Preservation of T cell Immune Response to the SARS-CoV-2 Omicron Variant]] | ||
|- valign="top" | |- valign="top" | ||
| '''authors''': | | '''authors''': | ||
| [[ | | [[Arnav Solanki]], [[Marc Riedel]], [https://orion.math.iastate.edu/cornette/ James Cornette], [[Julia Udell]], and [https://www.mayo.edu/research/faculty/vasmatzis-george-ph-d/bio-00027811 George Vasmatzis] | ||
|- valign="top" | |- valign="top" | ||
| ''' | | '''appeared in''': | ||
| [ | | [https://ieeexplore.ieee.org/document/10457533 IEEE/ACM Trans. on Computational Biology and Bioinformatics], pp. 1–11, March, 2024 | ||
|} | |} | ||
| | | width="70" align="center" | | ||
<span class="plainlinks"> | <span class="plainlinks"> | ||
[ | [https://mriedel.ece.umn.edu/wiki/images/1/12/Evasive_Spike_Variants_Elucidate_the_Preservation_of_T_Cell_Immune_Response_to_the_SARS-CoV-2_Omicron_Variant.pdf http://mriedel.ece.umn.edu/wiki/images/0/04/Pdf.jpg]</span> | ||
<br> | <br> | ||
[[Media: | [[Media:Evasive_Spike_Variants_Elucidate_the_Preservation_of_T_Cell_Immune_Response_to_the_SARS-CoV-2_Omicron_Variant.pdf | Paper]] | ||
|} | |} | ||
==Computing with Random Bit Streams== | |||
"''To invent, all you need is a pile of junk and a good imagination.''" –– '''Thomas A. Edison''' (1847–1931) | |||
Humans are accustomed to counting in a positional number system – [http://en.wikipedia.org/wiki/Decimal decimal] radix. Nearly all computer systems operate on another – [http://en.wikipedia.org/wiki/Binary_numeral_system binary] radix. We are so accustomed to these systems that it counterintuitive to ask: ''can we compute using a different representatio''n? and ''why would we want to''? | |||
== | ==== Stochastic Logic ==== | ||
We advocate an alternative representation: computing on random bit streams, where the signal value is encoded by the probability of obtaining a one versus a zero. Why compute this way? Using '''stochastic logic''', we can compute complex functions with very, very simple circuits. For instance, we can perform multiplication with a single AND gate and addition with a single MUX: | |||
{| align="center" | {| align="center" | ||
| | | [[Image:stochastic-multiplier.png|center|thumb|350px|'''Multiplication''' with an AND gate. Here the variables represents the ''probabilities'' of obtaining a 1 versus a 0 in stochastic bit streams. The AND gate produces an output probability <math>c</math> that is the product of the of the input probabilities <math>a</math> and <math>b</math>.]] | ||
[[Image: | || | ||
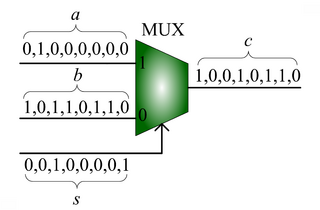
|| | || [[Image:stochastic-adder.png|thumb|320px|'''Scaled addition''' with a multiplexer (MUX). | ||
| Given input probabilities <math>a</math>, <math>b</math> and <math>s</math>, the MUX produces an output probability <math>c = a s + (1 - s) b</math>.]] | ||
|| | |} | ||
[[Image: | |||
|} | |||
Using conventional binary, building a circuit that computes, say a polynomial approximation to a trigonometric function such as ''tanh(x)'' or ''cos(x),'' requires ''thousands'' of logic gates. With stochastic logic, we have shown that we can compute such functions with about a ''dozen'' logic gates, so a '''100X reduction in gate count'''. Our most important contribution is a '''general methodology for synthesizing polynomial functions''' with stochastic logic, one of the seminal contributions to the field: | |||
{| | {| | ||
| | | | ||
{| style="background:#F0E68C" | {| style="background:#F0E68C" | ||
|- valign="top" | |- valign="top" | ||
| width="100" | '''title''': | | width="100" | '''title''': | ||
| width="500" | [ | | width="500" | [//mriedel.ece.umn.edu/wiki/images/2/21/Qian_Li_Riedel_Bazargan_Lilja_An_Architecture_for_Fault-Tolerant_Computation_with_Stochastic_Logic.pdf An Architecture for Fault-Tolerant Computation with Stochastic Logic] | ||
|- valign="top" | |- valign="top" | ||
| '''authors''': | | '''authors''': | ||
| [[ | | [[Weikang Qian]], [http://www.arctic.umn.edu/people.shtml Xin Li], [[Marc Riedel]], [http://www.ece.umn.edu/users/kia/ Kia Bazargan], and [http://www.arctic.umn.edu/lilja.shtml David Lilja] | ||
|- valign="top" | |- valign="top" | ||
| ''' | | '''appeared in''': | ||
| [http:// | | [http://ieeexplore.ieee.org/xpl/freeabs_all.jsp?arnumber=5601694 IEEE Transactions on Computers], Vol. 60, No. 1, pp. 93–105, 2011 | ||
|} | |} | ||
| | | width="70" align="center" | | ||
<span class="plainlinks">[http:// | <span class="plainlinks"> | ||
<br>[ | [http://cadbio.com/wiki/images/2/21/Qian_Li_Riedel_Bazargan_Lilja_An_Architecture_for_Fault-Tolerant_Computation_with_Stochastic_Logic.pdf http://mriedel.ece.umn.edu/wiki/images/0/04/Pdf.jpg]</span> | ||
<br> | |||
[//mriedel.ece.umn.edu/wiki/images/2/21/Qian_Li_Riedel_Bazargan_Lilja_An_Architecture_for_Fault-Tolerant_Computation_with_Stochastic_Logic.pdf Paper] | |||
|} | |} | ||
We have | ====Logic that Generates Probabilities==== | ||
We have also shown how to synthesize logic that transforms a set of ''source'' probabilities into different ''target'' probabilities. | |||
[[File:Generate_Probabilities_Example.png|center|frame|Given a set ''S'' of source probabilities {0.4, 0.5}, we can synthesize a combinational circuit to generate an arbitrary ''decimal'' output probability. The example shows how to generate 0.119. Each AND gate performs a multiplication and each inverter performs a "one-minus" operation.]] | |||
{| | {| | ||
| Line 171: | Line 262: | ||
|- valign="top" | |- valign="top" | ||
| width="100" | '''title''': | | width="100" | '''title''': | ||
| width="500" | [[Media: | | width="500" | [[Media:Qian_Riedel_Zhou_Bruck_Transforming_Probabilities_with_Combinational_Logic.pdf | Transforming Probabilities with Combinational Logic]] | ||
|- valign="top" | |- valign="top" | ||
| '''authors''': | | '''authors''': | ||
| [[ | | [[Weikang Qian]], [[Marc Riedel]], [http://paradise.caltech.edu/~hzhou/ Hongchao Zhou], and [http://paradise.caltech.edu/bruck.html Jehoshua Bruck] | ||
|- valign="top" | |- valign="top" | ||
| ''' | | '''will appear in''': | ||
| [http:// | | [http://tcad.polito.it/ IEEE Trans. on Computer-Aided Design of Integrated Circuits and Systems], 2012. | ||
|- valign="top" | |||
| '''presented at''': | |||
| [http://www.iccad.com/events/eventdetails.aspx?id=106-5-C International Conference on Computer-Aided Design], San Jose, 2009<br>(nominated for '''IEEE/ACM William J. McCalla ICCAD Best Paper Award'''). | |||
|} | |} | ||
| align="center" width="70" | | | align="center" width="70" | | ||
<span class="plainlinks"> | <span class="plainlinks"> | ||
[http:// | [http://cadbio.com/wiki/images/d/db/Qian_Riedel_Zhou_Bruck_Transforming_Probabilities_with_Combinational_Logic.pdf http://mriedel.ece.umn.edu/wiki/images/0/04/Pdf.jpg]</span> | ||
<br> [ | <br> | ||
[[Media:Qian_Riedel_Zhou_Bruck_Transforming_Probabilities_with_Combinational_Logic.pdf | Paper]] | |||
| align="center" width="70" | | | align="center" width="70" | | ||
<span class="plainlinks">[http://mriedel.ece.umn.edu/ | <span class="plainlinks">[http://mriedel.ece.umn.edu/files/Qian_Riedel_Bazargan_Lilja_The_Synthesis_of_Combinational_Logic_to_Generate_Probabilities.ppt http://mriedel.ece.umn.edu/wiki/images/3/36/Ppt.jpg]</span> | ||
<br> [http://mriedel.ece.umn.edu/ | <br> [http://mriedel.ece.umn.edu/files/Qian_Riedel_Bazargan_Lilja_The_Synthesis_of_Combinational_Logic_to_Generate_Probabilities.ppt Slides] | ||
|} | |} | ||
'''<big>A Deterministic Approach</big>''' | |||
Having championed stochastic logic for many years, we decided to reexamine its foundations. Why can complex functions be computed with such simple circuits when we compute on probabilities? Intuition might suggest that somehow we are harnessing deep aspects of probability theory. ''This intuition is <u>wrong</u>''. | |||
The keys is that we operate on uniform representation rather than a positional one. We showed that '''we can compute deterministically using the same structures that we use when computing stochastically.''' ''There is no need to do anything randomly!'' This upended the field that we had pioneered. | |||
{| | {| | ||
| Line 195: | Line 294: | ||
|- valign="top" | |- valign="top" | ||
| width="100" | '''title''': | | width="100" | '''title''': | ||
| width="500" | [ | | width="500" | [//mriedel.ece.umn.edu/wiki/images/c/c9/Najafi_Jenson_Lilja_Riedel_Performing_Stochastic_Computation_Deterministically.pdf Performing Stochastic Computation Deterministically] | ||
|- valign="top" | |- valign="top" | ||
| '''authors''': | | '''authors''': | ||
| [[ | | [[Devon Jenson]], [[M. Hassan Najafi]], [https://ece.umn.edu/directory/lilja-david/ David Lilja], and [[Marc Riedel]] | ||
|- valign="top" | |- valign="top" | ||
| '''appeared | | '''appeared in''': | ||
| [ | | [https://ieeexplore.ieee.org/document/8793244 IEEE Trans. on Very Large Scale Integration Systems],<br>Vol. 27, No. 29, pp. 2925–2938, 2019 | ||
|- valign="top" | |- valign="top" | ||
| '''presented at''': | | '''presented at''': | ||
| [ | | [https://iscas2020.org IEEE International Symposium of Circuits and Systems], 2020 | ||
|- valign="top" | |- valign="top" | ||
| '''presented at''': | | '''presented at''': | ||
| [http://www. | | [http://www.ieee.org/conferences_events/conferences/conferencedetails/index.html?Conf_ID=38629 IEEE/ACM International Conference on Computer-Aided Design], 2016 | ||
|} | |} | ||
| | | width="70" align="center" | | ||
<span class="plainlinks"> | <span class="plainlinks"> | ||
[http://mriedel.ece.umn.edu/wiki/images/ | [http://mriedel.ece.umn.edu/wiki/images/c/c9/Najafi_Jenson_Lilja_Riedel_Performing_Stochastic_Computation_Deterministically.pdf http://mriedel.ece.umn.edu/wiki/images/0/04/Pdf.jpg]</span> | ||
<br> | <br> | ||
[//mriedel.ece.umn.edu/wiki/images/c/c9/Najafi_Jenson_Lilja_Riedel_Performing_Stochastic_Computation_Deterministically.pdf Paper] | |||
| width="70" align="center" | | |||
| | <span class="plainlinks">[http://cadbio.com/wiki/images/4/4f/Jenson_Riedel_A_Deterministic_Approach_to_Stochastic_Computing.pptx http://mriedel.ece.umn.edu/wiki/images/3/36/Ppt.jpg]</span> | ||
[ | <br>[http://cadbio.com/wiki/images/4/4f/Jenson_Riedel_A_Deterministic_Approach_to_Stochastic_Computing.pptx Slides] | ||
[ | |||
|} | |} | ||
====Time-Encoded Computing==== | |||
Computing deterministically on bit streams really means that, instead of encoding data in '''''space''''', we encode them '''''time'''''. The time-encoding consists of periodic signals, with the value encoded as the fraction of the time that the signal is in the high (on) state compared to the low (off) state in each cycle. | |||
{| align="center" | {| align="center" | ||
| | |[[File:Analog-in-time.jpg|center|thumb|500px|Encoding a value in time. The value represented is the fraction of the time that the signal is high in each cycle, in this case 0.687.]] | ||
[[ | | | ||
| | ||[[File:Multiplicaiton-on-time-encoding-signals.jpg|thumb|500px|Multiplication with a single AND gate, operating on deterministic periodic signals. ]] | ||
|} | |||
|| | As technology has scaled and device sizes have gotten smaller, the supply voltages have dropped while the device speeds have improved. Control of the dynamic range in the voltage domain is limited; however, control of the length of pulses in the time domain can be precise. Encoding data in the time domain can be done more accurately and more efficiently than converting signals into binary radix. So we can compute ''more precisely,'' ''faster'', and with ''fewer logic gates:'' | ||
[[ | {| | ||
|- | |||
| | |||
{| style="background:#F0E68C" | |||
|- valign="top" | |||
| width="100" | '''title''': | |||
| width="500" | [//mriedel.ece.umn.edu/wiki/images/b/b0/Najafi_Jamali_Zavareh_Lilja_Riedel_Bazargan_Harjani_Time_Encoded_Values_for_Highly_Efficient_Stochastic_Circuits.pdf Time-Encoded Values for Highly Efficient Stochastic Circuits] | |||
|- valign="top" | |||
| '''authors''': | |||
| [[M. Hassan Najafi]], S. Jamali-Zavareh, [http://www.arctic.umn.edu/lilja.shtml David Lilja], [[Marc Riedel]], [http://people.ece.umn.edu/~kia Kia Bazargan] and<br>[http://people.ece.umn.edu/~harjani/ Ramesh Harjani] | |||
|- valign="top" | |||
| '''appeared in''': | |||
| [http://ieeexplore.ieee.org/xpl/aboutJournal.jsp?punumber=92 IEEE Trans. on Very Large Scale Integration Systems],<br>Vol. 25, No. 5, pp. 1644–1657, 2017 | |||
|- valign="top" | |||
| '''presented at''': | |||
| [http://www.ieee.org/conferences_events/conferences/conferencedetails/index.html?Conf_ID=33621 IEEE International Symposium on Circuits and Systems], 2017 | |||
|} | |} | ||
| width="70" align="center" | | |||
<span class="plainlinks"> | |||
[http://www.mriedel.ece.umn.edu/wiki/images/b/b0/Najafi_Jamali_Zavareh_Lilja_Riedel_Bazargan_Harjani_Time_Encoded_Values_for_Highly_Efficient_Stochastic_Circuits.pdf http://mriedel.ece.umn.edu/wiki/images/0/04/Pdf.jpg]</span> | |||
<br> | |||
[//mriedel.ece.umn.edu/wiki/images/b/b0/Najafi_Jamali_Zavareh_Lilja_Riedel_Bazargan_Harjani_Time_Encoded_Values_for_Highly_Efficient_Stochastic_Circuits.pdf Paper] | |||
|}Please see our "[[Papers,_Theses,_and_Presentations |Publications]]" page for more of our papers on these topics. | |||
==Computing with Feedback== | |||
"''A person with a new idea is a crank until the idea succeeds.''" –– '''Mark Twain''' (1835–1910) | |||
The accepted wisdom is that [http://en.wikipedia.org/wiki/Combinational_logic combinational circuits] (i.e., memoryless circuits) must have ''acyclic'' (i.e., loop-free or feed-forward) topologies. And yet simple examples suggest that this need not be so. We advocate the design of '''cyclic''' combinational circuits (i.e., circuits with loops or feedback paths). We have proposed a methodology for synthesizing such circuits and demonstrated that it produces significant improvements in area and in delay. | |||
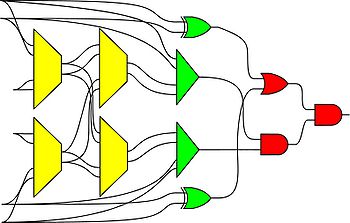
[[Image:cyclic-combinational-circuit.gif|center|thumb|none|550px|A circuit that has feedback and yet is combinational.]] | |||
{| | {| | ||
| | | | ||
| Line 263: | Line 363: | ||
|- valign="top" | |- valign="top" | ||
| width="100" | '''title''': | | width="100" | '''title''': | ||
| width="500" | [ | | width="500" | [[Media:Riedel_Bruck_Cyclic_Boolean_Circuits.pdf | Cyclic Boolean Circuits]] | ||
|- valign="top" | |- valign="top" | ||
| '''authors''': | | '''authors''': | ||
| [[ | | [[Marc Riedel]] and [http://paradise.caltech.edu/bruck.html Shuki Bruck] | ||
|- valign="top" | |||
| '''appeared in''': | |||
| [http://www.elsevier.com/wps/find/journaldescription.cws_home/505609/description#description Discrete Applied Mathematics], Vol. 160, No. 13–14, pp. 1877–1900, 2011. | |||
|- valign="top" | |||
| '''dissertation''': | |||
| Ph.D., [http://www.ee.caltech.edu Electrical Engineering], [http://www.caltech.edu Caltech], 2004<br>(winner of [http://www.ee2.caltech.edu/charles_wilts.html Charles H. Wilts Prize] for the '''Best Ph.D. Dissertation''' in EE at Caltech). | |||
|- valign="top" | |- valign="top" | ||
| '''presented at''': | | '''presented at''': | ||
| [http:// | | [http://www2.dac.com/40th/index.html Design Automation Conference], Anahiem, CA, 2003<br>(winner of '''DAC Best Paper Award'''). | ||
|} | |} | ||
| align="center" width="70" | | | align="center" width="70" | | ||
<span class="plainlinks"> | <span class="plainlinks"> | ||
[http://mriedel.ece.umn.edu/wiki/images/ | [http://mriedel.ece.umn.edu/wiki/images/9/94/Riedel_Bruck_Cyclic_Boolean_Circuits.pdf http://mriedel.ece.umn.edu/wiki/images/0/04/Pdf.jpg]</span> | ||
<br> | <br> | ||
[ | [[Media:Riedel_Bruck_Cyclic_Boolean_Circuits.pdf | Paper]] | ||
| align="center" width="70" | | | align="center" width="70" | | ||
<span class="plainlinks"> | <span class="plainlinks"> | ||
[http://mriedel.ece.umn.edu/wiki/images/ | [http://mriedel.ece.umn.edu/wiki/images/7/7a/Riedel_Cyclic_Combinational_Circuits.pdf http://mriedel.ece.umn.edu/wiki/images/0/04/Pdf.jpg]</span> | ||
<br> | <br> | ||
[http://mriedel.ece.umn.edu/wiki/images/ | [[Media:Riedel_Cyclic_Combinational_Circuits.pdf | PhD Dissertation]] | ||
| align="center" width="70" | | |||
<span class="plainlinks">[http://mriedel.ece.umn.edu/files/Riedel_Cyclic_Combinational_Circuits.ppt http://mriedel.ece.umn.edu/wiki/images/3/36/Ppt.jpg]</span> | |||
<br> [http://mriedel.ece.umn.edu/files/Riedel_Cyclic_Combinational_Circuits.ppt Slides] | |||
|} | |} | ||
Please see our [[Papers,_Theses,_and_Presentations | Publications]] page for more of our papers on this topic. | |||
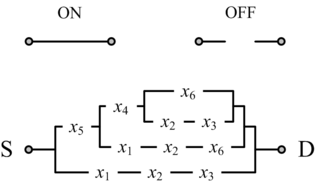
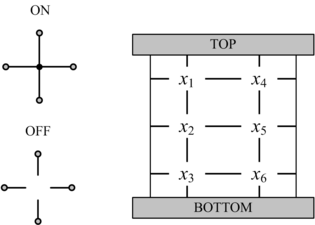
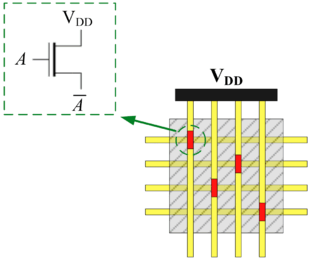
==Computing with Nanoscale Lattices== | ==Computing with Nanoscale Lattices== | ||
| Line 333: | Line 443: | ||
FET-like junctions that cross these develop a high or low impedance, respectively. | FET-like junctions that cross these develop a high or low impedance, respectively. | ||
]] | ]] | ||
|| | || | ||
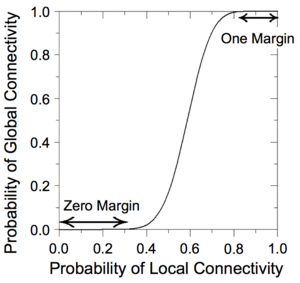
|| [[Image:percolation.gif|center|thumb| | || [[Image:percolation.gif|center|thumb|260.994x260.994px| In a switching network with defects, percolation can be exploited to produce robust Boolean functionality. ]] | ||
|} | |} | ||
We have devised a novel framework for digital computation with lattices of nanoscale switches with high defect rates, based on the mathematical phenomenon of [http://en.wikipedia.org/wiki/Percolation_theory percolation]. With random connectivity, percolation gives rise to a sharp non-linearity in the probability of global connectivity as a function of the probability of local connectivity. We exploit this phenomenon to compute Boolean functions robustly in the presence of defects. | We have devised a novel framework for digital computation with lattices of nanoscale switches with high defect rates, based on the mathematical phenomenon of [http://en.wikipedia.org/wiki/Percolation_theory percolation]. With random connectivity, percolation gives rise to a sharp non-linearity in the probability of global connectivity as a function of the probability of local connectivity. We exploit this phenomenon to compute Boolean functions robustly in the presence of defects. | ||
| Line 362: | Line 472: | ||
<br> [http://mriedel.ece.umn.edu/wiki/images/f/fe/Altun_Riedel_Neuhauser_Nanoscale_Digital_Computation_Through_Percolation.ppt Slides] | <br> [http://mriedel.ece.umn.edu/wiki/images/f/fe/Altun_Riedel_Neuhauser_Nanoscale_Digital_Computation_Through_Percolation.ppt Slides] | ||
|} | |} | ||
Please see our "[[Papers,_Theses,_and_Presentations |Publications]]" page for more of our papers on these topics. | Please see our "[[Papers,_Theses,_and_Presentations |Publications]]" page for more of our papers on these topics. | ||
==Algorithms and Data Structures== | ==Algorithms and Data Structures== | ||
| Line 414: | Line 478: | ||
"''There are two kinds of people in the world: those who divide the world into two kinds of people, and those who don't.''" –– '''Robert Charles Benchley''' (1889–1945) | "''There are two kinds of people in the world: those who divide the world into two kinds of people, and those who don't.''" –– '''Robert Charles Benchley''' (1889–1945) | ||
Consider the task of designing a digital circuit with 256 inputs. From a mathematical standpoint, such a circuit performs mappings from a space of <math>2^{256}</math> Boolean input values to Boolean output values. (The number of rows in a [http://en.wikipedia.org/wiki/Truth_table truth table] for such a function is approximately equal to [http://en.wikipedia.org/wiki/Observable_universe#Matter_content the number of atoms in the universe] – <math>10^{77}</math> rows versus <math>10^{79}</math> atoms!) Verifying such a function, let alone designing the corresponding circuit, would seem to be an intractable problem. Circuit designers have succeeded in their endeavor largely as a result of innovations in the data structures and algorithms used to represent and manipulate [http://en.wikipedia.org/wiki/Boolean_function Boolean functions]. We have developed novel, efficient techniques for synthesizing functional dependencies based on so-called [http://en.wikipedia.org/wiki/Boolean_satisfiability_problem SAT-solving algorithms]. We use [http://en.wikipedia.org/wiki/Craig_interpolation Craig Interpolation] to generate circuits from the corresponding Boolean functions. | Consider the task of designing a digital circuit with 256 inputs. From a mathematical standpoint, such a circuit performs mappings from a space of <math>2^{256}</math> Boolean input values to Boolean output values. (The number of rows in a [http://en.wikipedia.org/wiki/Truth_table truth table] for such a function is approximately equal to [http://en.wikipedia.org/wiki/Observable_universe#Matter_content the number of atoms in the universe] – <math>10^{77}</math> rows versus <math>10^{79}</math> atoms!) Verifying such a function, let alone designing the corresponding circuit, would seem to be an intractable problem. | ||
Circuit designers have succeeded in their endeavor largely as a result of innovations in the data structures and algorithms used to represent and manipulate [http://en.wikipedia.org/wiki/Boolean_function Boolean functions]. We have developed novel, efficient techniques for synthesizing functional dependencies based on so-called [http://en.wikipedia.org/wiki/Boolean_satisfiability_problem SAT-solving algorithms]. We use [http://en.wikipedia.org/wiki/Craig_interpolation Craig Interpolation] to generate circuits from the corresponding Boolean functions. | |||
{| align="center" | {| align="center" | ||
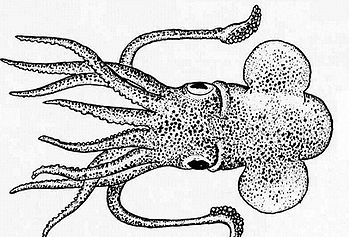
| [[Image:sat-squid.jpg|center|thumb|350px|A circuit construct for SAT-based verification.]] | | [[Image:sat-squid.jpg|center|thumb|350px|A circuit construct for SAT-based verification.]] | ||
| Line 445: | Line 511: | ||
"''Mathematics may be defined as the subject in which we never know what we are talking about, nor whether what we are saying is true.''" –– '''Bertrand Russell''' (1872–1970) | "''Mathematics may be defined as the subject in which we never know what we are talking about, nor whether what we are saying is true.''" –– '''Bertrand Russell''' (1872–1970) | ||
[[Image:xkcd_disciplines_by_purity.png|center|thumb|740x740px]] | |||
The great mathematician [http://en.wikipedia.org/wiki/John_von_Neumann John von Neumann] articulated the view that research should never meander too far down theoretical paths; it should always be guided by potential applications. This view was not based on concerns about the relevance of his profession; rather, in his judgment, real-world applications give rise to the most ''interesting'' problems for mathematicians to tackle. At their core, most of our research contributions are mathematical contributions. The tools of our trade are [http://en.wikipedia.org/wiki/Discrete_mathematics discrete math], including [http://en.wikipedia.org/wiki/Combinatorics combinatorics] and [http://en.wikipedia.org/wiki/Probability probability theory]. | The great mathematician [http://en.wikipedia.org/wiki/John_von_Neumann John von Neumann] articulated the view that research should never meander too far down theoretical paths; it should always be guided by potential applications. This view was not based on concerns about the relevance of his profession; rather, in his judgment, real-world applications give rise to the most ''interesting'' problems for mathematicians to tackle. At their core, most of our research contributions are mathematical contributions. The tools of our trade are [http://en.wikipedia.org/wiki/Discrete_mathematics discrete math], including [http://en.wikipedia.org/wiki/Combinatorics combinatorics] and [http://en.wikipedia.org/wiki/Probability probability theory]. | ||
{| | {| | ||
| | | | ||
Latest revision as of 15:31, 24 July 2025
"You see things; and you say, 'Why?' But I dream things that never were; and I say, 'Why not?'"
–– George Bernard Shaw (1856 –1950)
Our research spans different disciplines ranging from digital circuit design, to algorithms, to mathematics, to synthetic biology. It tends to be inductive (as opposed to deductive) and conceptual (as opposed to applied). A recurring theme is building systems that compute in novel or unexpected ways with new and emerging technologies.
Storing Data with Molecules
All new ideas pass through three stages:
- It can't be done.
- It probably can be done, but it's not worth doing.
- I knew it was a good idea all along!
––Arthur C. Clarke (1917–2008)
Ever since Watson and Crick first described the molecular structure of DNA, its information-bearing potential has been apparent. With each nucleotide in the sequence drawn from the four-valued alphabet of {A, T , C, G}, a molecule of DNA with n nucleotides stores 2n bits of data.
- Could we store data for our computer systems in DNA? "Can't be done – too hard."
- Is it worth doing? "Definitely not. It will never work as well as our hard drives do."
- But one can store so much data so efficiently! "I knew it was a good idea all along!"
|

|

|
Computing with Molecules
"Biology is the most powerful technology ever created. DNA is software, protein are hardware, cells are factories."
––Arvind Gupta (1953– )
Computing has escaped! It has gone from desktops and data centers into the wild. Embedded microcontrollers – found in our gadgets, our buildings, and even our bodies – are transforming our lives. And yet, there are limits to where silicon can go and where it can compute effectively. It is a foreign object that requires a electrical power source.
We are studying novel types of computing systems that are not foreign, but rather an integral part of their physical and chemical environments: systems that compute directly with molecules. A simple but radical idea: compute with acids and bases. An acidic solution corresponds to a "1" and a basic solution to "0".
|
It's more complex that acids and bases, but DNA is a terrific chassis for computing. We have developed "CS 101" algorithms with DNA: Sorting, Shifting and Searching:
|
Based on a bistable mechanism for representing bits, we have implemented logic gates such AND, OR, and XOR gates, as well as sequential components such as latches and flip-flops with DNA. Using these components, we have built full-fledged digital circuits such as a binary counters and linear feedback shift registers.
|
Also, we have performed signal processing including operations such as filtering and fast-fourier transforms (FFTs) with DNA.
|
Please see our "Publications" page for more of our papers on these topics.
Computational Immunology
“Physics is the study of the simple things in the Universe. Biology is the study of the complex ones. ”
–– Richard Dawkins (1941– )
We are studying a problem that computer science currently judges to be very difficult: predicting cellular immunity. It centers on the question of how strongly molecules binds to one another. The molecules in question are peptides – fragments of proteins from a virus – binding to cell-surface receptors. A peptide will only bind if it fits like a key into a lock.
The binding is a critical step in a critical component of the immune system: it allows circulating T-cells to kill off infected cells. If this mechanism succeeds, an infection is stopped in its tracks. If it fails, then infected cells become factories for reproducing copies of the virus; full-blown disease results. Given a novel pathogen, such as SARS-Cov-2, predicting whether the immune system of an individual will do its job at fighting off the disease comes down to predicting how well the viral peptides bind to the cell-surface receptors of that person. We are tackling the problem with cloud computing resources, donated by Oracle.
|
|
Computing with Random Bit Streams
"To invent, all you need is a pile of junk and a good imagination." –– Thomas A. Edison (1847–1931)
Humans are accustomed to counting in a positional number system – decimal radix. Nearly all computer systems operate on another – binary radix. We are so accustomed to these systems that it counterintuitive to ask: can we compute using a different representation? and why would we want to?
Stochastic Logic
We advocate an alternative representation: computing on random bit streams, where the signal value is encoded by the probability of obtaining a one versus a zero. Why compute this way? Using stochastic logic, we can compute complex functions with very, very simple circuits. For instance, we can perform multiplication with a single AND gate and addition with a single MUX:
Using conventional binary, building a circuit that computes, say a polynomial approximation to a trigonometric function such as tanh(x) or cos(x), requires thousands of logic gates. With stochastic logic, we have shown that we can compute such functions with about a dozen logic gates, so a 100X reduction in gate count. Our most important contribution is a general methodology for synthesizing polynomial functions with stochastic logic, one of the seminal contributions to the field:
|
Logic that Generates Probabilities
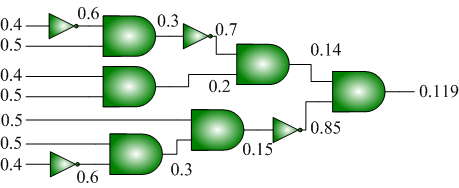
We have also shown how to synthesize logic that transforms a set of source probabilities into different target probabilities.
|
A Deterministic Approach
Having championed stochastic logic for many years, we decided to reexamine its foundations. Why can complex functions be computed with such simple circuits when we compute on probabilities? Intuition might suggest that somehow we are harnessing deep aspects of probability theory. This intuition is wrong.
The keys is that we operate on uniform representation rather than a positional one. We showed that we can compute deterministically using the same structures that we use when computing stochastically. There is no need to do anything randomly! This upended the field that we had pioneered.
|
Time-Encoded Computing
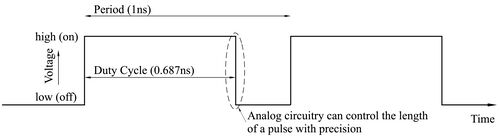
Computing deterministically on bit streams really means that, instead of encoding data in space, we encode them time. The time-encoding consists of periodic signals, with the value encoded as the fraction of the time that the signal is in the high (on) state compared to the low (off) state in each cycle.
As technology has scaled and device sizes have gotten smaller, the supply voltages have dropped while the device speeds have improved. Control of the dynamic range in the voltage domain is limited; however, control of the length of pulses in the time domain can be precise. Encoding data in the time domain can be done more accurately and more efficiently than converting signals into binary radix. So we can compute more precisely, faster, and with fewer logic gates:
|
Please see our "Publications" page for more of our papers on these topics.
Computing with Feedback
"A person with a new idea is a crank until the idea succeeds." –– Mark Twain (1835–1910)
The accepted wisdom is that combinational circuits (i.e., memoryless circuits) must have acyclic (i.e., loop-free or feed-forward) topologies. And yet simple examples suggest that this need not be so. We advocate the design of cyclic combinational circuits (i.e., circuits with loops or feedback paths). We have proposed a methodology for synthesizing such circuits and demonstrated that it produces significant improvements in area and in delay.
|
Please see our Publications page for more of our papers on this topic.
Computing with Nanoscale Lattices
"Listen to the technology; find out what it’s telling you.” –– Carver Mead (1934– )
In his seminal Master's Thesis, Claude Shannon made the connection between Boolean algebra and switching circuits. He considered two-terminal switches corresponding to electromagnetic relays. A Boolean function can be implemented in terms of connectivity across a network of switches, often arranged in a series/parallel configuration. We have developed a method for synthesizing Boolean functions with networks of four-terminal switches. Our model is applicable for variety of nanoscale technologies, such as nanowire crossbar arrays, as molecular switch-based structures.
|
The impetus for nanowire-based technology is the potential density, scalability and manufacturability. Many other novel and emerging technologies fit the general model of four-terminal switches. For instance, researchers are investigating spin waves. A common feature of many emerging technologies for switching networks is that they exhibit high defect rates.
We have devised a novel framework for digital computation with lattices of nanoscale switches with high defect rates, based on the mathematical phenomenon of percolation. With random connectivity, percolation gives rise to a sharp non-linearity in the probability of global connectivity as a function of the probability of local connectivity. We exploit this phenomenon to compute Boolean functions robustly in the presence of defects.
|
Please see our "Publications" page for more of our papers on these topics.
Algorithms and Data Structures
"There are two kinds of people in the world: those who divide the world into two kinds of people, and those who don't." –– Robert Charles Benchley (1889–1945)
Consider the task of designing a digital circuit with 256 inputs. From a mathematical standpoint, such a circuit performs mappings from a space of Boolean input values to Boolean output values. (The number of rows in a truth table for such a function is approximately equal to the number of atoms in the universe – rows versus atoms!) Verifying such a function, let alone designing the corresponding circuit, would seem to be an intractable problem.
Circuit designers have succeeded in their endeavor largely as a result of innovations in the data structures and algorithms used to represent and manipulate Boolean functions. We have developed novel, efficient techniques for synthesizing functional dependencies based on so-called SAT-solving algorithms. We use Craig Interpolation to generate circuits from the corresponding Boolean functions.
|
Please see our "Publications" page for more of our papers on this topic. (Papers on SAT-based circuit verification, that is, not on squids.)
Mathematics
"Mathematics may be defined as the subject in which we never know what we are talking about, nor whether what we are saying is true." –– Bertrand Russell (1872–1970)
The great mathematician John von Neumann articulated the view that research should never meander too far down theoretical paths; it should always be guided by potential applications. This view was not based on concerns about the relevance of his profession; rather, in his judgment, real-world applications give rise to the most interesting problems for mathematicians to tackle. At their core, most of our research contributions are mathematical contributions. The tools of our trade are discrete math, including combinatorics and probability theory.
|
Please see our "Publications" page for more of our papers on this topic.

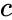 that is the product of the of the input probabilities
that is the product of the of the input probabilities 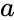 and
and 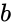 .
.
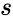 , the MUX produces an output probability
, the MUX produces an output probability 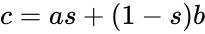 .
.
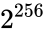 Boolean input values to Boolean output values. (The number of rows in a
Boolean input values to Boolean output values. (The number of rows in a 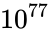 rows versus
rows versus 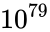 atoms!) Verifying such a function, let alone designing the corresponding circuit, would seem to be an intractable problem.
atoms!) Verifying such a function, let alone designing the corresponding circuit, would seem to be an intractable problem.